Auto Annotation
Have you ever been sent a plain unannotated sequence, or downloaded a sequence from Entrez and been disappointed as it doesn't have the carefully curated graphical appearance of your favorite genes? Auto annotation solves both of these common problems. The basic idea is that you can scan the sequence against a folder containing a collection of existing annotated sequences and MacVector will find the matchingfeatures in the folder and add those to the starting sequence.
Annotating a Vector
Many cloning vectors are posted online as plain sequences with a corresponding PDF showing a map of the features on the vector. Normally, the only way to create an annotated version of a vector like this is to laboriously enter each individual feature into a feature editor. Now, you can speed up the process dramatically by using the auto-annotation function to quickly annotate common features. If the vector has additional features not present in your existing sequence collection, you can then manually add those features, save the new vector in your collection, and the next time you come across a similar vector, auto annotation will automaticaly add the new features.
pET104-DEST
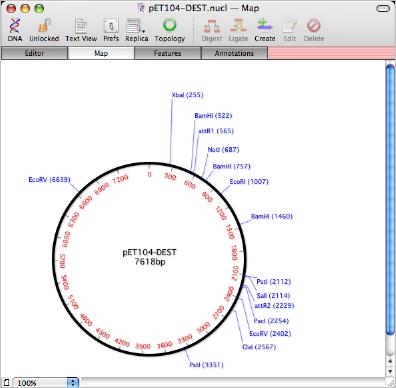
This is a Gateway cloning vector from Invitrogen. You can find the Invitrogen supplied map here. The sequence you can download from Invitrogen has no annotations, so when opened in MacVector the Map tab shows a plain circle.
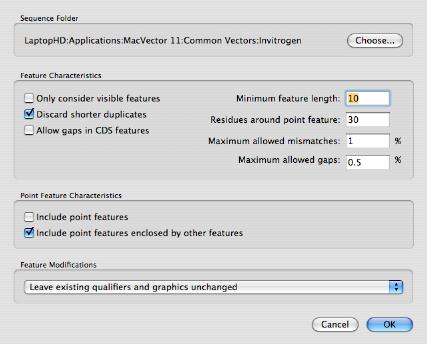
You can then use the menu item Database | Auto-annotate Sequence... to bring up the auto-annotation configuration dialog. Here, the target folder has been set to the /Common Vectors/Invitrogen/ folder that gets installed with MacVector 11 and contains a variety of vectors.
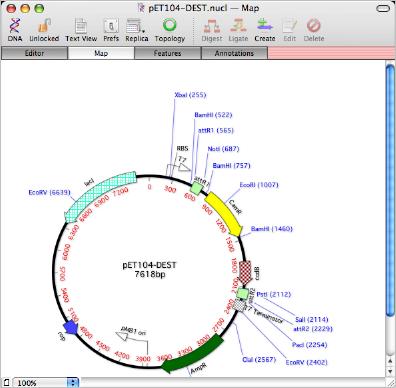
After clicking OK, the folder is scanned and and the vector gets updated with matching features from the folder. Note how each feature is given the graphical appearance of the feature as it existed in the target folder.
|

2x.png)


2x.png)
