Align to Reference - Sequence Confirmation
MacVector has a unique Align to Reference interface that lets you align one or more files against a reference sequence. There are two main uses for this: Sequence Confirmation is similar to sequence assembly, except that it requires the use of a known reference sequence as a scaffold. The second use is cDNA Alignment, which allows you to align mRNA, cDNA or EST sequences against a genomic template.
You can use this functionality to help you solve a number of typical laboratory problems:
- Confirming the sequence of a cloned fragment
- Sequencing across the ends of a cloned fragment to confirm the junction sequence
- Screening clones from a site-specific mutagenesis experiment to identify successful mutations
- Screening related clones for single nucleotide polymorphisms
The main limitation of this implementation is that you must have a reference sequence to act as a scaffold against which the sample sequences can be aligned. You therefore cannot use this to assemble trace files from de novo sequencing projects. To do that, you should purchase the optional MacVector Assembler add-on module that uses the industry standard phred and phrap algorithms from the University of Washington.
There is a detailed Sequence Confirmation Tutorial that provides far more information on this functionality that can be downloaded here. The trial version of MacVector includes all of the sample files you will need to follow the tutorial.
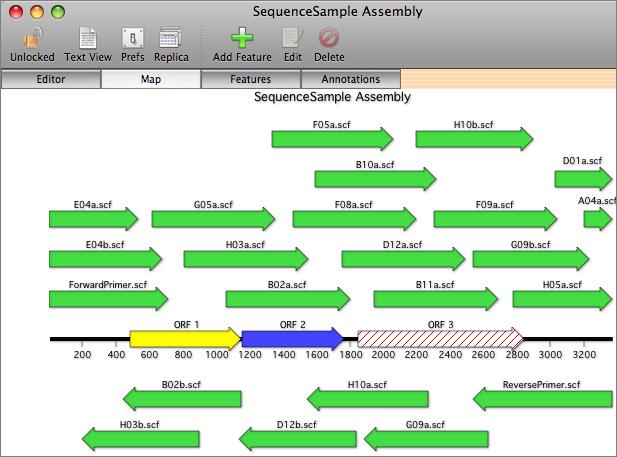
Sequence Confirmation Window
The key to this functionality lies in the main Sequence Confirmation Window. You start with a reference sequence - this can be the predicted sequence that you are trying to confirm, or a reference clone if you are screening for SNPs or mutagenesis results. This is displayed along the top of the window. You can then add one or more automatic sequencing trace files to the window by clicking on the "+" button. The imported sequences (MacVector supports ABI, SCF and ALF formats) are displayed below the reference sequence and the associated traces are displayed in a lower graphical pane. Initially, the sequence residues are displayed in italics to indicate they have not been aligned.
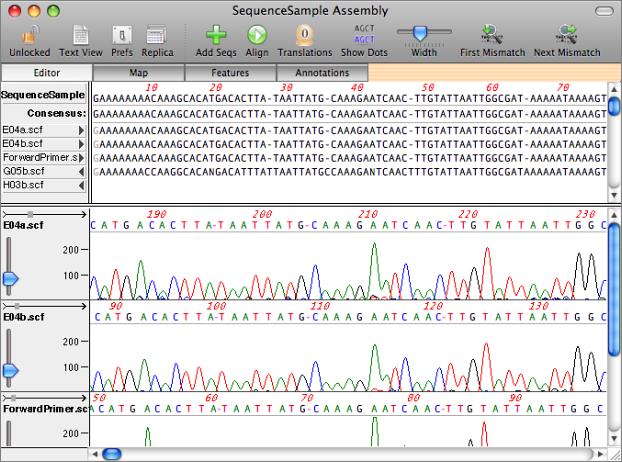

Aligning Imported Sequences
The sequences can be aligned by clicking on the alignment button (the circular arrow). MacVector uses a custom algorithm that is fine tuned to align closely related sequences that may have many short insertions/deletions. Once aligned, MacVector provides a number of tools to help you identify mismatches between the sample sequences and the reference sequence. Clicking on the reference or consensus sequence aligns all of the traces in the center of the display so that you can directly compare the raw data at that position. A search tool quickly locates positions where the consensus of the sample sequence differs from the reference and the residues can be viewed with dots substituted wherever residues match the reference. The "dots" mode is particularly effective for identifying SNPs and other single nucleotide differences between sequences.

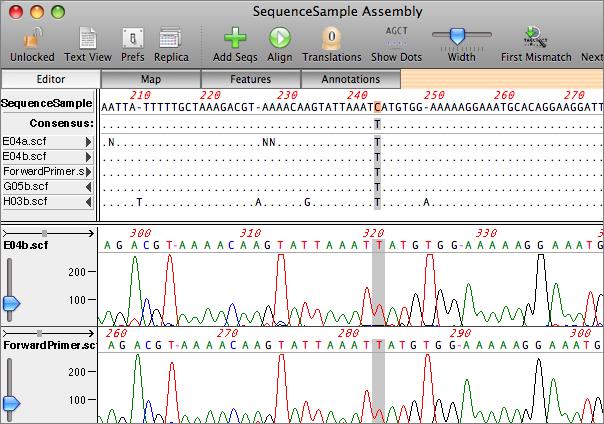

|

2x.png)


2x.png)
