Assembling Sequences Using phrap
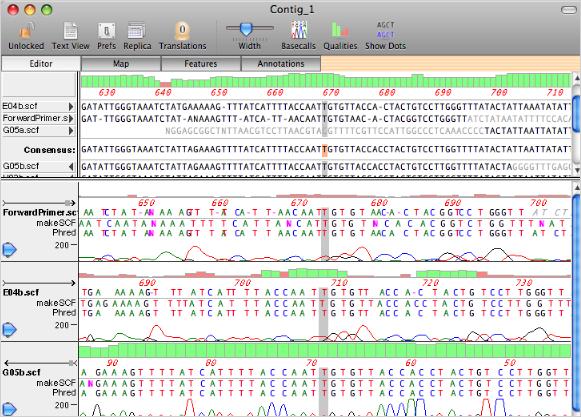
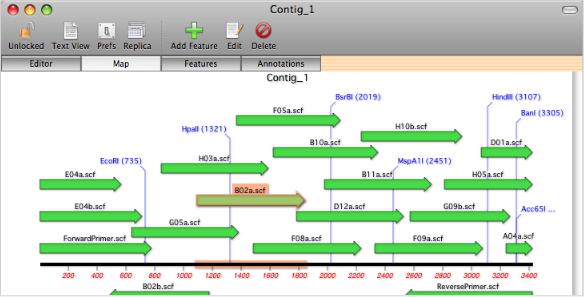
MacVector Assembler assembles sequences using the phrap algorithm. phrap does not require the sequences to have been base called by phred, or to have had any vector sequences masked. However, using phred and cross_match will improve the accuracy of phrap assemblies. phrap assembles sequences into contigs and creates a consensus sequence with its own set of quality values, based on the quality and strandedness of the overlapping sequences. Contigs can be viewed and edited in a contig editor that shows the aligned sequences along with the chromatograms in a lower pane. Clicking on a residue in the consensus sequence resets the chromatogram view so that they are all aligned to that base. This allows you to easily align the chromatograms so that you can resolve ambiguities in the consensus sequence.
|
2x.png)

2x.png)
