MacVector 18.8 is out and it’s packed with new tools! MacVector 18.8 has tools to help you identify and annotate unknown, unannotated or partially annotated sequences. Ideal for identifying contigs from a de novo assembly. One of these new tools is AutoAnnotate (via BLAST)
Auto-Annotate (via BLAST) is similar to Auto-Annotate (local), except instead of using curated sequences on your own Mac (for example the Annotated fragments folder) it uses BLAST to find similar sequences in the NCBI’s BLAST database then annotates your sequence with matching features found in those hits.
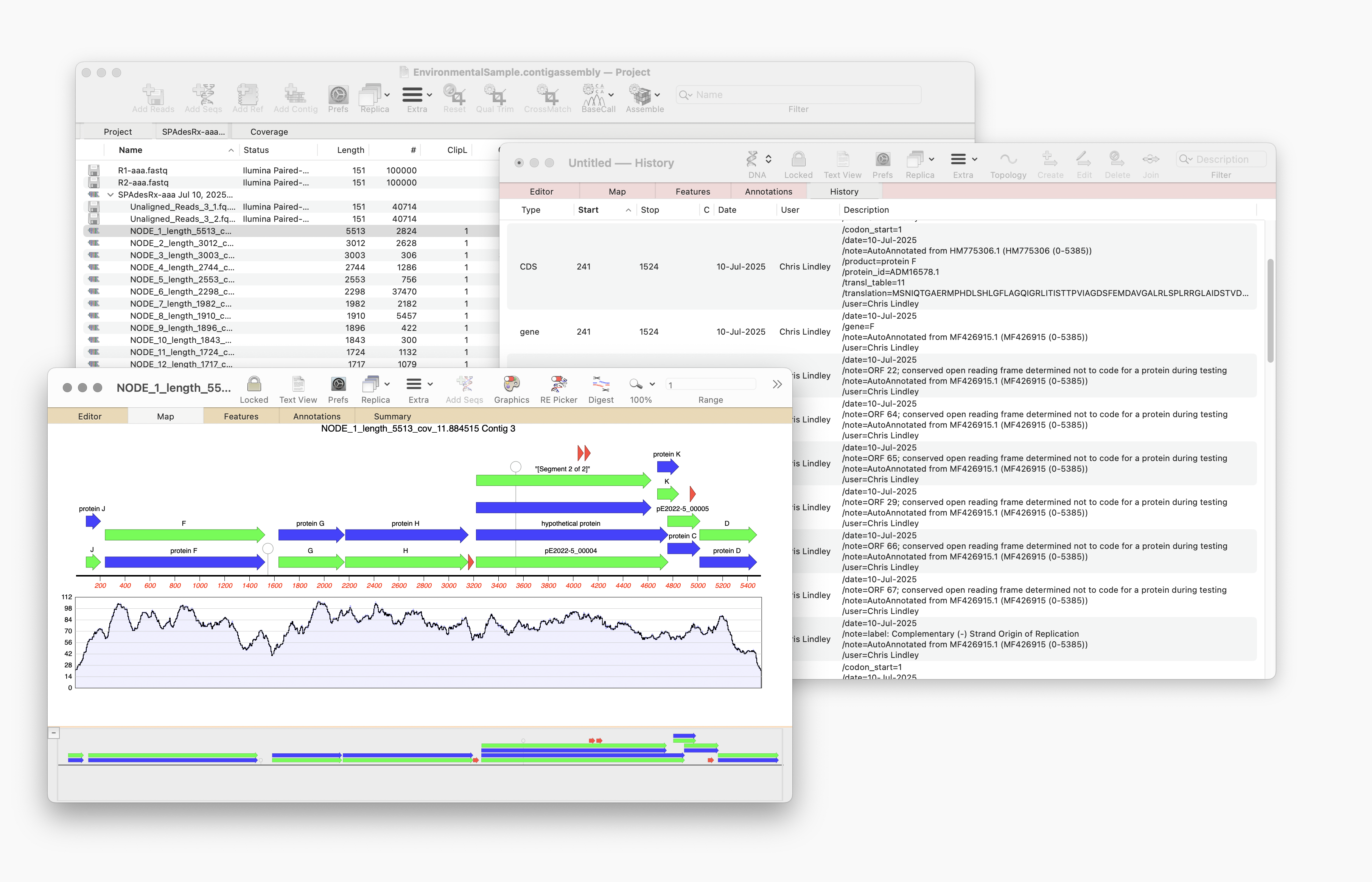
Auto-Annotate (via BLAST) will run on any nucleic acid sequence. However, where it is particularly useful is running directly from the Assembly Project manager to identify contigs that you have assembled. Just select one or hundreds of contigs and run. Every contig will be annotated with features and will aid in identification.
How to automate sequence annotation
- Open an existing Assembly Project with contigs you want to identify.
- go to FILE | NEW | ASSEMBLY PROJECT
- Click ADD SEQS and add all your query sequences.
- Select all your query sequences
- Choose DATABASE | AUTO ANNOTATE SEQUENCE (VIA BLAST)…
- The upper pane of the setup dialog shows options about the BLAST search.
- The lower pane shows options about what features to annotate. If you wish to modify existing features to change their graphical attributes, change the drop down menu to Replace Features and Qualifiers for Existing Features. If you wish to just create new features then select Leave Existing Features and Qualifiers Unchanged. You can also just modify existing features by selecting Replace only Graphics for Existing Features.
- Click OK.
For longer or many sequences searches will take a considerable time. However, you can close the job and allow it to be run in the background using the Job Manager.
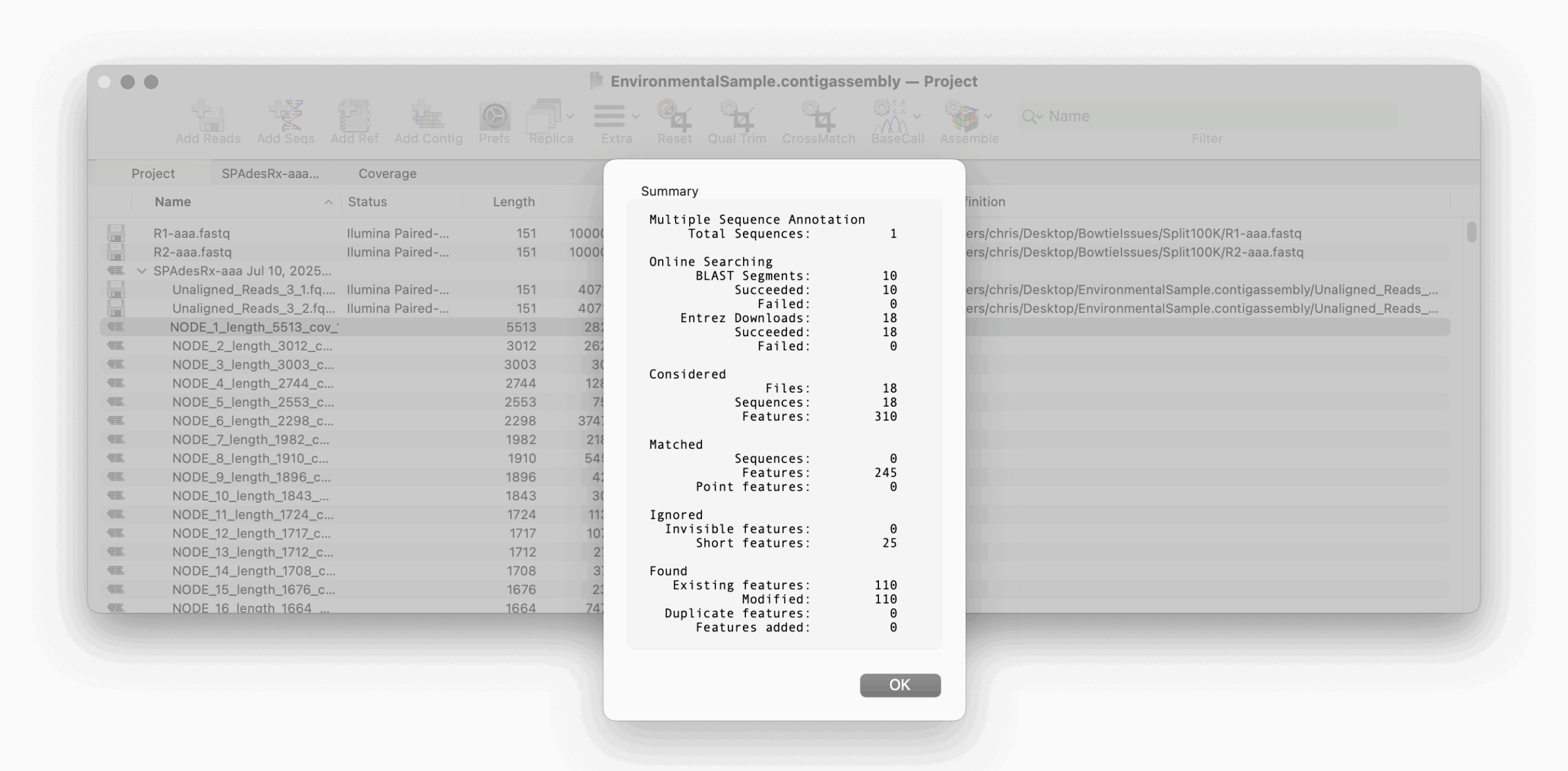
The results will be displayed in a Summary dialog, then annotated directly onto the sequence(s).
