What's New in MacVector 13?
Overview
MacVector 13 has had a significant interface overhaul, with new icons, redesigned tab controls, tabbed result windows with tear-off tabs and enhanced graphical resolution for retina displays. The Quicktest and Primer3 design tools have been enhanced to simplify the design and testing of primers with mismatches, including the display of restriction enzyme sites around the binding location and identification of one-out sites with silent mutations in overlapping open reading frames. The Assembler module has been enhanced to include the popular Velvet de novo assembly algorithm for NGS data along with changes to the phrap algorithm to better assemble both traditional ABI chromatogram files and NGS data. Plus, of course, there's a slew of minor enhancements aimed at simplifying workflows through MacVector. A full list of the changes can be found in the Release Notes. There have also been a series of blog posts on the new features in MacVector 13, discussing the changes to the Primer Design functionality, changes to the result windows, the addition of the popular Velvet de novo assembly algorithm and an overview of the interface changes.
Interface Enhancements
All of the toolbar icons have been reworked to have a more consistent look and feel and to display at high resolution on Retina screens. The tab bars have been redesigned on all of the primary windows.
All result windows now get collated into a single result window per sequence, with tabs for each result. If you want to view more than one result at a time, you can “tear off” any tab into its own window.
There is a new File | Export Tab Contents menu item that can be used to save tab- specific data (e.g. graphics or text).
Primer Design Enhancements
The Quicktest Primer interface now shows restrictions enzyme cut sites in the parental sequence around the primer binding site and also shows “one out” restriction enzyme sites. These are colored according to the effect that changing the mismatched base would have on anyoverlapping annotated ORFs . Potential silent mutations, which would not change the amino acid sequence of any coding sequences, are shown in red, those that would change the code are shown in green.
Hovering over a restriction site with the mouse displays the actual recognition sequence of the site. Clicking and holding on a site, temporarily changes the sequence of the primer to match the mutated site and changes to the amino acid sequence of any ORFs are displayed above the sequence. Double-clicking on a one-out site changes the primer sequence to match the selected site. This can be seen graphically in the following animation;
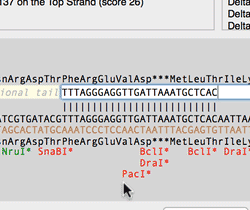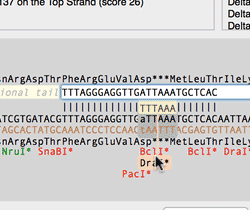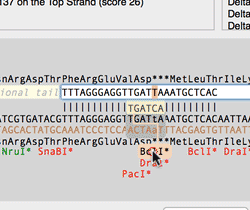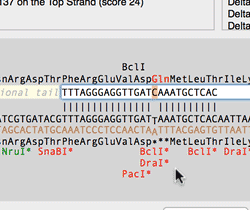
The Primer3 interface has been enhanced for testing pairs of primers. There is now a fourth “testing” mode that is automatically invoked when a pair of primers are entered. This mode removes most of the constraints to ensure pairs of binding primers are correctly identified. The optimal annealing temperature for pairs of primers is now displayed in the spreadsheet result. Primer3 will now include primers that overlap the ends of the region to be amplified. Previously, primers had to be entirely outside of the region.
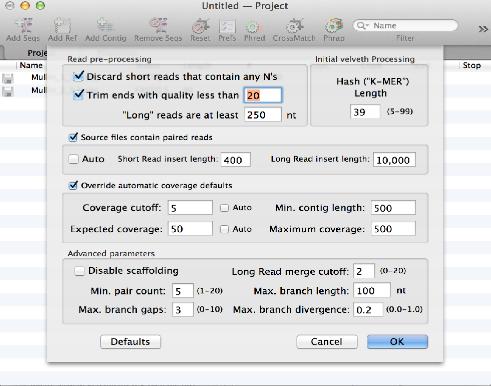
Assembling de novo NGS Sequences With Velvet
The popular Velvet algorithm has been added to the Assembly module. You can use this to assemble large numbers of NGS sequences supplied in fastq files. In our hands, around 20 million 90nt Illumina reads can be assembled into a 6 Mb bacterial genome in around an hour on a retina MacBook Pro with 16 GB RAM.
Applescript Support
Some basic Applescript capabilities have been added to MacVector 13. In particular, there are now commands to open and save files in specified formats, so you can e.g. create Applescripts that can iterate through a folder of sequences and save each of them into a different folder in any format supported by MacVector. There is a new Applescripts folder with some example scripts to get you started.
Chromatogram Files
There is a new Raw Data tab that lists the peak positions, basecalls, quality values and areas under the curve for the chromatogram trace data. There are also columns that indicate the potential presence of “mixed” residues at a variety of confidence levels. It is in a tab-delimited format that can be copied and pasted directly into Excel for additional analysis.
Miscellaneous Changes
The Starting Point dialog now has links to the Entrez browser and to help pages to help new users understand the many different ways you can get sequences into MacVector.
The Align To Folder function now reads fastq formatted files in the target folder. The Description result window now displays the full name of each sequence, even with the typically long names from NGS experiments.
The phrap default parameters have been tweaked to generate better assemblies of both NGS and chromatogram data.
All of the “plain text” views now automatically adjust their formatting when printed to ensure that they are no more than a single page in width.
The Page Preview and Magnify controls have been moved from the bottom left corner of map windows onto the toolbar.
|

2x.png)


2x.png)
